Chapter 7.7: Reproducible Research and Workflow Organization
This chapter examines systematic workflow organization principles that enable reproducible research through structured design patterns, parameterization strategies, and maintenance protocols. Key concepts include modular workflow architecture, configuration management approaches, error handling mechanisms, and collaborative framework implementation that support scientific validity and research transparency across distributed academic environments.
Foundations of Reproducible Research Design
Reproducible research represents a research methodology that enables different researchers to obtain identical results from the same data through standardized procedures, transparent methodology, and comprehensive documentation. This approach requires systematic workflow design that separates data input, transformation logic, and output generation into discrete components that can be independently verified and validated across different computational environments and research teams.
The COVID-19 Research Database initiative at Johns Hopkins University School of Medicine exemplifies systematic workflow organization in large-scale collaborative research. Dr. Casey Overby Taylor’s team coordinated longitudinal health data analysis across 15 medical centers involving over 40 researchers who needed to process identical analytical procedures on different institutional datasets while maintaining scientific reproducibility standards. The research program required systematic tracking of patient outcomes, symptom progression, and treatment effectiveness across diverse populations over multiple years.
KNIME Analytics Platform’s workflow architecture supports reproducible research through modular design patterns that organize analytical components by function including data preparation modules, analytical procedures, quality assurance checks, and output formatting sections. This systematic organization enables research teams to understand and modify specific elements without affecting overall workflow integrity while maintaining the analytical transparency essential for peer review and regulatory compliance.
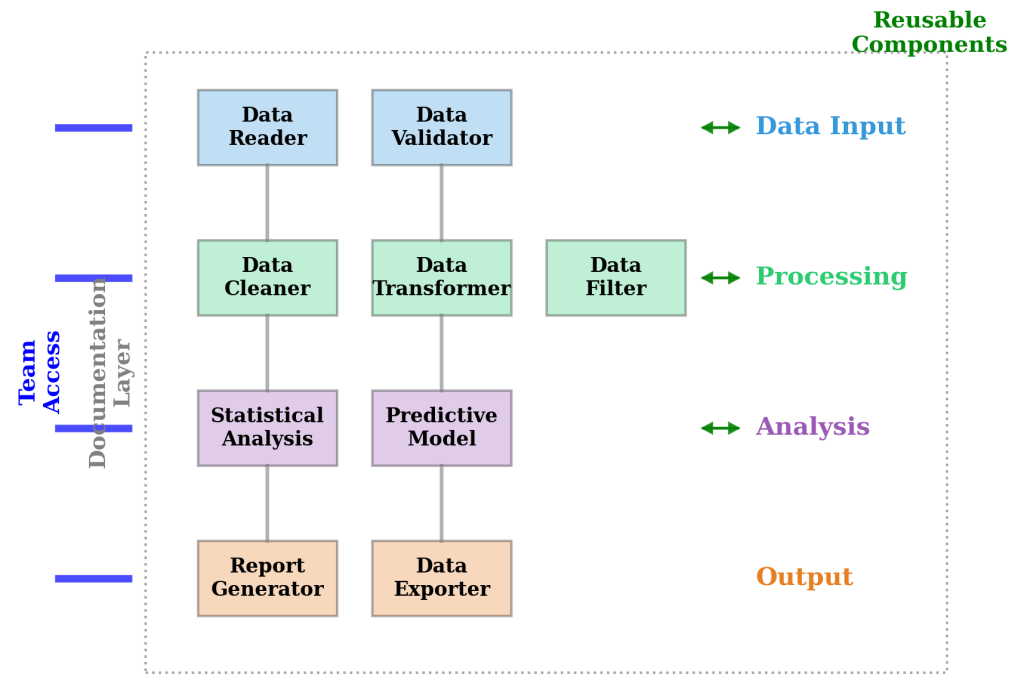
Figure 7.7.1: Systematic workflow organization showing hierarchical structure from data input through processing, analysis, and output stages. The modular design demonstrates clear component separation with documentation layers, collaboration access points, and quality assurance checkpoints integrated throughout the analytical pipeline.
Systematic Folder Organization: Implement standardized metanode templates that encapsulate common analytical procedures including data cleaning, statistical analysis, and visualization generation. Create naming conventions for workflow components using formats such as “DataCleaning_PatientOutcomes_v2.1_2024-08-15” that incorporate functional descriptions, version information, and modification dates enabling research teams to track analytical evolution and identify current procedures.
Documentation standards must capture analytical rationale, parameter settings, validation procedures, and expected outputs in ways that enable independent researchers to replicate findings without consulting original developers. These standards include data lineage tracking that identifies all source systems, transformation procedures, and quality assurance steps supporting academic publication and research funding requirements.
Parameterization and Configuration Management Strategies
Workflow parameterization transforms static analytical procedures into flexible frameworks that adapt to different datasets, research contexts, and analytical requirements through systematic configuration management. This approach involves implementing Configuration Nodes that expose critical parameters including file paths, filtering criteria, statistical thresholds, and output formats through user-friendly interfaces that enable researchers to execute workflows with appropriate settings for their specific datasets.
KNIME’s Configuration Nodes and Workflow Variables enable systematic modification without workflow restructuring by creating global parameters such as study period dates, institutional identifiers, and analytical categories that automatically propagate throughout complex workflows. This approach ensures consistency across all analytical components while reducing manual configuration errors that could compromise research validity.
Parameter validation procedures verify configuration settings before workflow execution, preventing errors that could compromise research results or waste computational resources. Effective parameterization includes automated testing mechanisms that confirm parameter combinations produce expected results across different datasets while maintaining analytical validity required for peer-reviewed publication.
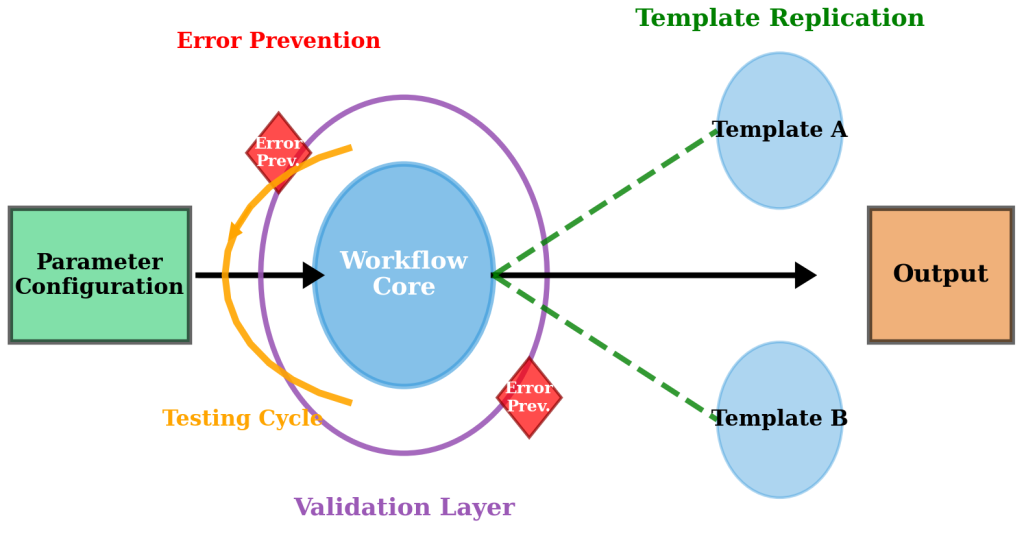
Figure 7.7.2: Configuration management framework showing central workflow core with parameter inputs, validation controls, and template replication capabilities. The framework demonstrates systematic parameter testing cycles and error prevention mechanisms that enable workflow adaptation across different research contexts.
Parameter template development involves creating validated starting points for common research scenarios including longitudinal studies, cross-sectional analyses, and comparative effectiveness research. These templates provide standardized configurations while maintaining flexibility for study-specific modifications through systematic parameter governance procedures that track configuration changes and maintain audit trails of analytical decisions.
Error Handling and Maintenance Protocols
Robust error handling ensures workflow reliability across varying data conditions through systematic exception management, validation procedures, and recovery mechanisms that prevent analytical failures from compromising research timelines. This approach involves implementing Try-Catch structures that identify data quality issues, missing files, and processing errors while providing informative messages that enable researchers to diagnose and resolve problems independently. Validation checkpoints throughout workflows verify data integrity, parameter settings, and intermediate results at critical processing stages, ensuring errors are detected early rather than propagating through complex analytical procedures. Fallback procedures implement alternative processing paths when primary analytical approaches encounter unexpected data conditions, maintaining workflow execution continuity while preserving analytical validity through documented exception handling protocols.
The Johns Hopkins COVID-19 research consortium implemented comprehensive error detection that identified data quality issues including missing demographic information, inconsistent coding systems, and temporal data gaps across participating institutions. Error handling procedures provided standardized reporting formats that enabled consistent interpretation and resolution across different institutional contexts while maintaining research timeline integrity.
Maintenance protocols support long-term workflow sustainability through systematic testing procedures, update management, and version control that accommodate evolving research requirements and technological changes. Automated testing workflows validate analytical procedures against reference datasets, ensuring modifications do not introduce errors or change established results while supporting continuous improvement initiatives.
Systematic Monitoring Implementation: Establish monitoring systems that track workflow performance, identify processing bottlenecks, and alert research teams to potential issues before they affect critical research deadlines. These capabilities include resource utilization tracking that optimizes computational efficiency while maintaining analytical accuracy across different institutional computing environments and varying dataset scales.
Change management procedures balance research agility with analytical stability through systematic impact assessment, stakeholder notification, and rollback capabilities that preserve research continuity. Documentation standards track all workflow modifications, testing results, and validation procedures supporting continuous quality improvement while maintaining analytical transparency essential for reproducible research.
Collaborative Framework Implementation
Collaborative reproducible research requires organizational frameworks that enable multiple researchers to contribute to analytical development while maintaining consistency, quality, and scientific validity. These frameworks incorporate role-based access controls that define contributor responsibilities, review procedures that ensure analytical accuracy, and communication protocols that coordinate development activities across distributed research teams. Standardized development environments ensure analytical consistency across different institutional computing resources through systematic software version management, dependency tracking, and computational environment documentation. These standards enable research teams to replicate analytical environments and achieve consistent results regardless of local hardware specifications or institutional computing policies.
Knowledge transfer protocols support research continuity through comprehensive documentation that captures analytical rationale, implementation decisions, and validation procedures in formats that enable effective handoffs between research team members. These protocols include troubleshooting guides and best practices documentation that accelerate new team member integration while preserving institutional analytical knowledge.
The KNIME workflow library developed by Johns Hopkins has been published through the KNIME Community Hub and serves as a template for other longitudinal health research initiatives. The systematic organization approach enabled the research consortium to publish findings in high-impact journals including The New England Journal of Medicine and Nature Medicine, with complete reproducibility documentation that allows other research teams to replicate and extend the analytical methods.
Integration with Research Communication
Reproducible workflow organization directly supports research communication requirements by creating systematic documentation frameworks that enable clear presentation of analytical methods to diverse stakeholder audiences. Well-organized workflows facilitate the translation of complex analytical procedures into comprehensible methodological descriptions suitable for academic publication, regulatory submission, and stakeholder reporting. The systematic organization principles developed through collaborative research initiatives continue to support ongoing investigations and have been adapted for multi-site longitudinal studies in cardiovascular disease and cancer research. These frameworks demonstrate how reproducible research practices enable knowledge transfer across research domains while maintaining methodological rigor and supporting the communication transparency essential for scientific advancement.
Systematic workflow organization transforms ad hoc analytical processes into scalable frameworks that meet academic research standards while supporting professional communication requirements across diverse research environments and stakeholder contexts.

